The idea of using bacterial cells for storing information has been thought of for a long time. This idea was a result of the large quantity of new information rapidly generated on a daily basis [1]. At present, the International Data Corporation (IDC) estimated that on a daily basis, about 2.5 quintillion bytes of data are generated, much of which is machine and human-generated data, and the pace is rapidly accelerating. The IDC further estimated that by 2025, newly generated data up to 180 zettabytes (ZB) orbytes 1.8 x1023 bytes will be created. These figures are very large to fathom. To perfectly understand these large estimates, the estimates exceed the radius of the observable universe which is 43 x 1021 km, exactly two orders of magnitude lesser than the said estimate of bytes [2].
The use of machine learning techniques has further intensified this need for an alternative means of storing a large volume of information. This technique entails the use of social networks, and navigation systems that run over big volumes of data. To end this problem of data storage, alternative means of information storage are required. One promising solution is the use of bacterial Deoxyribonucleic acid (DNA). This storage technique uses non-motile bacteria and the information to be stored is digitally encoded. The whole process works by the use of bacterial Nano networks which allow for molecular communication [3].
As technological advancement continues to move in a rapid manner, as evident by the high technology computers and phones we use, which come with high resolution cameras, that continue to consume much space, there is the fear that if an alternative as well as a sustainable means of data storage is not devised, the breakthrough in the technological industries will be halted, and this might affect the computer/electronic industries as well as global economies. This challenge has made researchers dwell into finding alternative ways of data storage, which entails the use of bacterial cells. This might not be in connection with the obvious fact that bacteria retain the credit of one of the most abundant life forms on earth with the ability to grow into millions within just 18 to 24 hours.
Of all the bacterial cells on earth amounting to bacterial taxonomists argued that only about less than 5% have been discovered. This 5% is thought to reach hundreds of thousands of different bacteria each with unique distinguishing properties the diverse groups of bacteria, the bacteria Escherichia coli, usually abbreviated as E. coli has proven to be the most promising candidate in DNA Molecular Data Storage Systems. The bacteria (E. coli) remains the only bacteria whose genome (a pool containing all the genetic elements of a cell) has largely been studied, this allows for the modification via the alteration of given cell properties. This often results in many breakthroughs in the field of Microbiology, from gene therapy, production of recombinant vaccines and single-cell proteins, and Bacterial intelligence, a technique that integrates cell Programmeming with genetic circuit which allows for the E. coli cell to solve the diverse classical problem in artificial intelligence that today are solved by computers [3], and many more techniques to be available in the near future.
The bacterial DNA encodes all the genetic information encoded by the cell which comprises all the cell characteristics and is capable of replicating in other for the cell to maintain its generation. This is achieved as the cell continues to grow. In bacterial cells, growth is defined as an increase in the number of cells as opposed to an increase in size. This storage technique offers numerous advantages, as the copy of stored information increases as the bacterial cells grow. This generates many different copies of a single data that is stored, hence decreasing the likelihood of missing information. More so, this storage technique offers the opportunity to store condensed information with long-term stability. Before now, the main challenge faced by Molecular Microbiologists and other Bioinformaticians is the means by which this information can be safely retrieved. However, subsequent studies on Microbial genetics and molecular biology approach were able to provide solutions to these problems as scientists were able to find a more promising technique of data retrieval of all stored information via the use of genetically engineered motile bacteria, which handle the task of reading operations. Although reading operations has been identified as a solution to the problem, the challenge now is how the entire reading process of the encoded information can be automated, especially if the stored information is archived all together. Recent developments in bacterial studies suggest the use of molecular communications which uses the cell molecular signaling mechanisms via a quorum sensing approach. In this approach, the bacterial DNA is seen as the software whereas the whole cell is seen as the hardware component.
The whole idea of the use of bacteria as storage devices (Biostorage) employs the use of bacterial nanonetworks (figure 1) along with some plasmids. The plasmids are extra chromosomal material containing some set of information, that confer on bacteria some given properties. In this technique, the plasmids are encoded with some set of needed digital information usually from an archive library and the engineered bacteria (E. Coli) are made to pick up those plasmids. The plasmids will then deliver the encoded information into the cell for sequencing operations and then decode the information. Simulations (imitation of the operation of a given system) are then employed to assess how the information is transferred into the cell as well as assess how it affects the E. coli cells as it is assembled towards a given destination in the cell.
To assess the ability of the bacteria to pick the data of choice for storage, as well as the retrieval of the archived information, techniques such as; Molecular Positioning System, digitally encoded-DNA archive system, System validation, and Wet lab experiments (figure 2 and 3) must be taken into consideration.
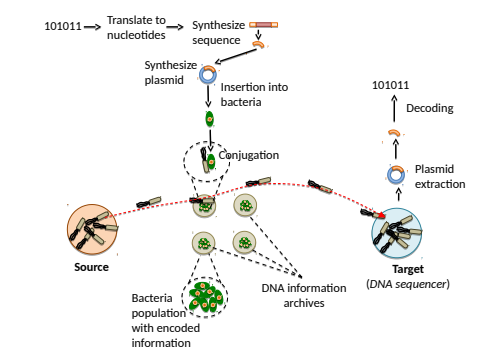
Figure 1: Overall system architecture of the DNA archival system that enables reading using bacterial nanonetworks. Motile bacteria (in grey) swim from the Source towards each of the archive points, which contain different information. Reached the motile-restricted bacteria, conjugation starts to retrieve the encoded information contained into the plasmid. Last step, motile bacteria swim towards the Target, where the plasmid is retrieved through sequencing processes [4].
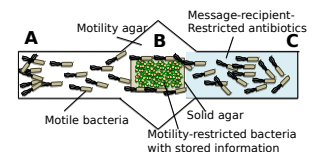
Figure 2: Illustration of the wet lab experimental setup, where the motility-restricted bacteria with the encoded “Hello World” is placed at B. Non-motile bacteria swim from A towards C and pick up the message along the way [4].
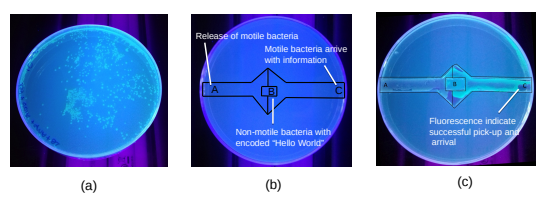
Figure 3: Wet lab experimental result demonstrating the successful pick-up of the encoded “Hello World” message from the non-motile bacteria in B by the motile bacteria released at A. After picking up the message, the motile bacteria swims towards C to deliver the message. (a) demonstrates the successful conjugation process, (b) presents the engineered agar plate illustrating the placement of the non-motile bacteria with the encoded plasmid of “Hello World”, and the channel for the motile bacteria to swim from A to C, (c) results of the experiment, where the fluorescence indicates the motile bacteria successfully conjugated with the non-motile bacteria to pick up the encoded plasmid with antibiotic resistance gene and surviving the antibiotics [4].
Reference
[1] | A. Anzel, D. Heider and G. Hattab, “The visual story of data storage: From storage properties to user interfaces,” Computational and structural biotechnology , vol. 19, pp. 4904 – 4918, 2021. |
[2] | G. J., “Quantum fluctuations and the slow accelerating expansion of the the universe.,” EPL (Europhysics Letters) ;:. , no. 50002, p. 125, 2019. |
[3] | R. Lahoz-Beltra, J. Navarro and P. C. Marijuán, “Bacterial computing: a form of natural computing and its applications,” Frontiers in Microbiology, vol. 5, no. 1, p. 101, 2014 . |
[4] | F. Tavella, A. Giaretta, T. M. Dooley-Cullinane, M. Conti, L. Coffey and S. Balasubramaniam, “DNA Molecular Storage System: Transferring Digitally Encoded Information through Bacterial Nanonetworks,” The University of Manchester Research Explorer. ArXiv, vol. 1, no. 1, 2018. |
Ismail Rabiu is an Assistant Lecturer in the Department of Microbiology, Skyline University Nigeria (SUN). He holds an M.Sc in Medical Microbiology with a specialization in Pharmaceutical Microbiology and Infectious diseases.
You can join the conversation on facebook @SkylineUniversityNG and on twitter @SkylineUNigeria
